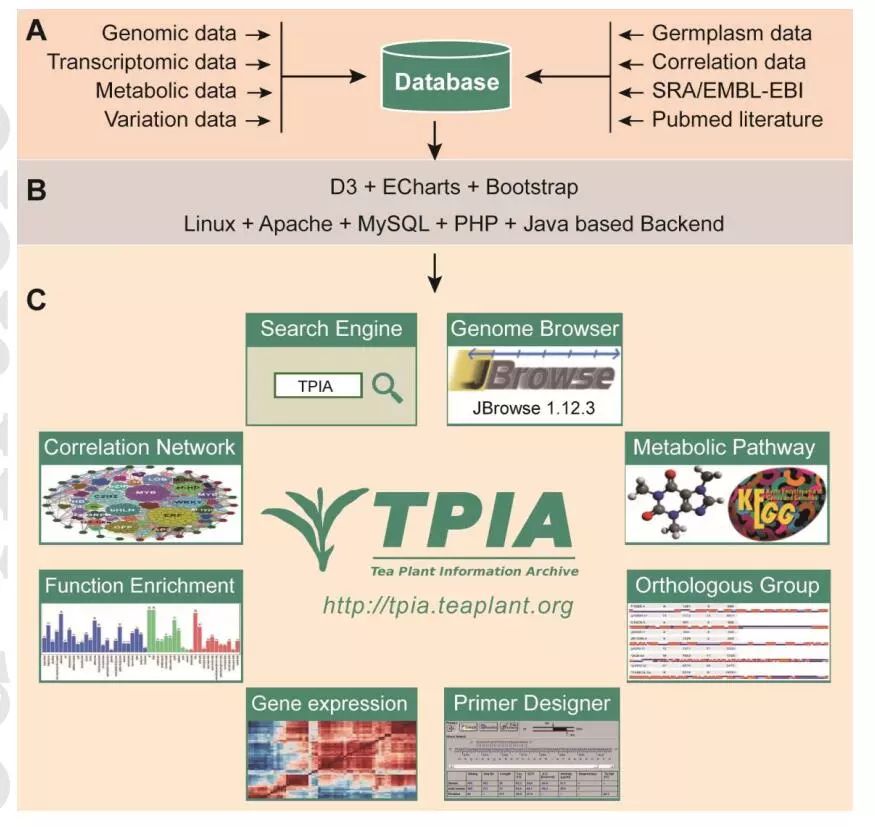
作物基因组学的发展在有效利用现代分子生物学进行作物遗传改良方面发挥了关键作用。尤其是近十年来在水稻、小麦、玉米等一般作物中,基于测序或者重测序技术从根本上促进了与重要农艺性状相关的大量基因的大规模克隆和鉴定,极大的加快了高产优质新品种的培育。而对于茶树,由于长期缺乏参考基因组,极大的限制了现代分子育种技术在茶树新品种选育中的应用。为了解决上述问题,安徽农大历时十余年完成了中国种茶树栽培种的基因组测序工作,公布了一个相对高质量的茶树参考基因组。而随着基因组测序技术的发展,大规模的组学数据呈现海量式增长,如何有效地整合和共享这些海量的数据,进而加快解决当前茶产业相关的重大科学问题,又成为了一个新的主要难题。现在,安徽农大的研究人员又基于‘舒茶早’的基因组数据、广泛整合现有的茶树转录组、代谢组以及种质资源等数据建立了第一个动态的、交互式的茶树基因组数据库:Tea Plant Information Archive (TPIA)。
该数据库包含:
1. 基因组数据
'舒茶早' 的3.14Gb (其中2.89Gb无缺失)基因组数据;33932个高质量编码基因;20个完整的BAC克隆和738个BAC末端序列;涵盖64%茶树无间隔基因组序列的1.86Gb TE序列;2486个TF;59765个SSR;阿萨姆种和中国种茶树基因组的151个共线区间;107个功能实验验证的基因信息。
2. 转录组数据
茶树8个组织的94.11GbRNA-seq数据;33932个茶树蛋白编码基因的表达数据;茶树冷胁迫、干旱胁迫、盐胁迫、MeSA处理的RNA-seq数据;9个茶树近源种、13个茶组植物种的转录组数据。
3. 代谢物数据
不同茶树组织、不同茶组植物的儿茶素类、原花青素、茶氨酸、咖啡碱代谢物含量数据。
4. 相关性数据
33932个茶树基因在不同组织、不同逆境处理下的基因共表达数据;基因表达与代谢物积累相关性数据。
5. 其他数据
SNP数据;DNA甲基化数据;非编码RNA数据;多于1100份的茶树种质信息。
上述数据均可下载:http://tpia.teaplant.org/download.html.
此外,该平台还提供了大量分析工具,比如基因组查看、基因搜索、基因分析、功能富集、相关分析等,并能对分析结果可视化。
Abstract
Tea is the world's widely consumed non‐alcohol beverage with essential economic and health benefits. Confronted with the increasing large‐scale omics dataset particularly the genome sequence released in tea plant, the construction of a comprehensive knowledgebase is urgently needed to facilitate the utilization of these datasets towards molecular breeding. We hereby present the first integrative and specially designed web‐accessible database, Tea Plant Information Archive (TPIA; http://tpia.teaplant.org). The current release of TPIA employs the comprehensively annotated tea plant genome as framework, and incorporates with abundant well‐organized transcriptomes, gene expressions (across species, tissues, and stresses), orthologs, and characteristic metabolites determining tea quality. It also hosts massive transcription factors, polymorphic simple sequence repeats, single nucleotide polymorphisms, correlations, manually curated functional genes, and globally collected germplasm information. A variety of versatile analytic tools (e.g. JBrowse, blast, enrichment analysis, etc.) are established helping users to perform further comparative, evolutionary, and functional analysis. We show a case application of TPIA that provides novel and interesting insights into the phytochemical content variation of section Thea of genus Camellia under a well‐resolved phylogenetic framework. The constructed knowledgebase of tea plant will serve as a central gateway for global tea community to better understand the tea plant biology that largely benefits the whole tea industry.
责任编辑:千鹤茶苗